This is an automatically translated article.
The article is written by Master, Doctor Mai Vien Phuong - Gastroenterologist - Department of Medical Examination & Internal Medicine - Vinmec Central Park International General Hospital.It has been noted that the diseased esophagus associated with healthy controls is home to a bacterial population unusually rich in gram-negative species. Insights into the esophageal microbiome could help guide further analysis of new therapeutic tools.
1. Esophageal microbiota is being studied extensively
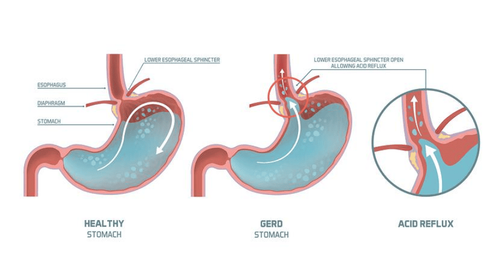
Surveying the gut microbiome has gradually changed the understanding of esophageal disease. Over the past two decades, 16S rRNA gene sequencing has been used to characterize and compare the esophageal microbiota of healthy individuals with those of patients with esophageal diseases, including gastroesophageal reflux disease. esophagitis (reflux esophagitis), Barrett's esophagus (BE), esophageal adenocarcinoma (EAC), eosinophilic esophagitis (EoE), and esophageal motility disorders.
Analysis of the compositional differences between healthy microorganisms and dysbiotics in the esophagus has provided more detailed information on the pathogenesis of reflux esophagitis, its sequelae, and associated complications. other related diseases. In particular, it has been noted that the diseased esophagus, associated with healthy controls, is home to a bacterial population unusually rich in gram-negative species. Furthermore, to a large extent, the bacteria are unsuitable for the pathogens that cause oral inflammation. Insights in the esophageal microbiome may help guide further investigation of new therapeutic tools targeting these mechanisms.
2. Changes in flora
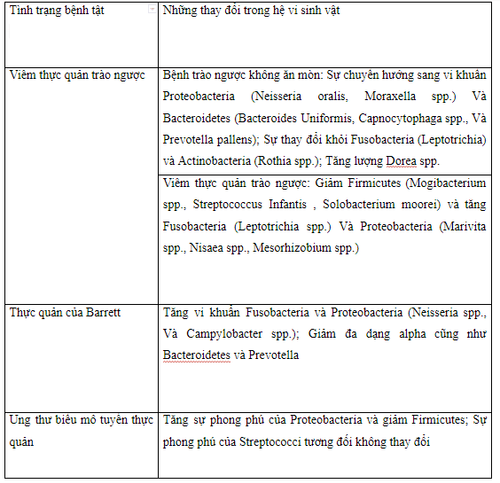
3. Characteristics of 'normal' and 'abnormal' esophageal microbiota
Genetic sequencing of the human population shows that the gastroesophageal microbiota is dominated by almost in order of prevalence by 6 major phyla: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria and Saccharibacteria. Normally, Bacteroidetes and Firmicute are dominant mainly in response to an abundance of Bacteroides or Clostridium spp. Here, phylum-level classification is an oversimplification that does not explain the diversity that exists in a relatively simple microbiome as found in the distal esophagus. Substantial differences in both identity and relative abundance of specific bacteria exist, particularly when taxa are characterized with higher resolution by species elucidation. or line level.
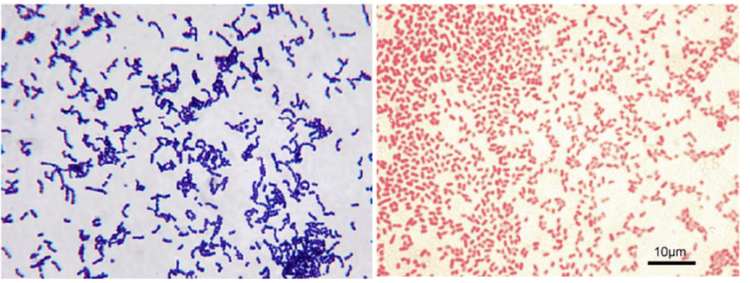
In 2009, a team identified two different types of esophageal microbiota. Comparing outcomes from individuals with reflux esophagitis with healthy controls, they characterized the control population, mainly gram-positive organisms, Streptococcus spp., as a microbiome. Type I organisms. Class II microbiota are largely associated with pathological conditions, such as reflux esophagitis, Barrett's esophagus, and show a relative increase in the number of anaerobic bacteria. gram negative.
4. Three distinct clusters of esophageal microbiota in esophageal tissue
Further studies identified the taxa and observed that three distinct clusters of esophageal microbiota dominated biopsies of human esophageal tissue. Each is characterized by relative abundance of Streptococcus and Prevotella spp.. Cluster 1 is an intermediate group, with approximately equal proportions of both genera with an increasing presence of Haemophilus and Rothia spp. Cluster 2 mainly consisted of Streptococcus spp. Cluster 3 is mainly represented by Prevotella spp.5. The composition of the gastrointestinal tract microbiota varies from mouth to stomach
In addition to inter-individual variability, the composition of the gastrointestinal microbiota also varies in the esophagus from the mouth to the stomach in both health and disease. Specifically, the combined flora of the proximal, middle, and distal esophagus varies both in composition as well as in relative abundance. In a study of 12 patients routinely monitored for Barrett's esophagus, the proximal pharynx was more similar to the pharynx in that it had a higher concentration of gram-positive microorganisms than the distal esophagus. Streptococcus spp. was found throughout the esophagus, with a relatively large increase from the proximal to the mid-oesophageal and a marked decrease thereafter in the distal esophagus.
Gram-negative organisms including Prevotella and Delftia spp., are generally more concentrated in the distal esophagus. This is not surprising, because the lipopolysaccharide (LPS) "shell" around gram-negative organisms hardens them with changes in pH, bile salt concentrations, proteases, and to some extent That's the temperature. This is why most enteric pathogens are gram-negative - they can survive selection and possible adverse effects related to the proximity to substances in the stomach. thick. Therefore, it can be expected that the microbiome becomes much less diverse and increasingly rich in gram-negative species.
6. The greatest diversity is in the area near the nasal cavity
Diversity is greatest near the nasal cavity and with mildest conditions, e.g. salivary/mucus, light physiological pH and moderate physiological temperature. One would expect to see a slope towards the prolific and obligate anaerobic bacteria (possibly from the subchondral space) in the lumen of the esophagus distal to the stomach.
Notably, many bacteria also have increased abundance because of the protective nature of spores (Clostridia spp.) in the harsh surroundings. Accordingly, the underlying esophageal microbiota appears to be related to changes in the composition of this continuum and the gram-positive/negative balance.
7. Biomic differences for esophageal disease
The biomic difference for esophageal disease is remarkable. Patients with Barrett's esophagus had higher overall levels of Streptococcus spp in whole-line tissue biopsies than those without the disease. Although this appears to contradict the gram-positive/negative imbalance discussed earlier, this may be an indirect effect of persistent local irritation due to immunogenic gram-negative species, generating conditions for bacteria to multiply and invade the underlying tissue with gram-positive bacteria, of which streptococci are a major part.
8. Conclusion
These findings suggest that the relative abundance of gram-negative flora in the distal esophagus leads to a local inflammatory response, which negatively impacts barrier function. This eventually leads to tissue proliferation of all flora, including Streptococcus spp. The proximal-distant variation in the specific flora changing from the healthy to the diseased state, combined with the esophageal microbiome subtype has previously illustrated the pathological potential of dysbiosis. Emerging data may provide insight into new therapeutic models, focusing on microbial alterations that may complement, even replace, the need for acid suppression in relevant states. to this disease.
Please dial HOTLINE for more information or register for an appointment HERE. Download MyVinmec app to make appointments faster and to manage your bookings easily.